Automatically plots the molecule with a camera position and field of view that includes the full model. For more control over the scene, pass the scene to `rayrender::render_scene()` or `rayvertex::rasterize_scene()` and specify the camera position manually. Note: spheres and cylinders in the scene are used to automatically compute the field of view of the scene–if rendering with rayrender, adding additional sphere (e.g. with `rayrender::generate_ground()`) will change this calculation. Use `rayrender::render_scene()` instead if this is a problem.
Arguments
- scene
`rayrender` scene of molecule model.
- fov
Default `NULL`, automatically calculated. Camera field of view.
- angle
Default `c(0,0,0)`. Degrees to rotate the model around the X, Y, and Z axes. If this is a single number, it will be taken as the Y axis rotation.
- order_rotation
Default `c(1,2,3)`. What order to apply the rotations specified in `angle`.
- lights
Default `top`. If `none`, removes all lights. If `bottom`, lights scene with light underneath model. If `both`, adds lights both above and below model. This can also be a matrix of light information generated with `rayvertex`.
- lightintensity
Default `80`. Light intensity for pathtraced scenes.
- ...
Other arguments to pass to `rayrender::render_scene()` or `rayvertex::rasterize_scene()`
Value
Rendered image
Examples
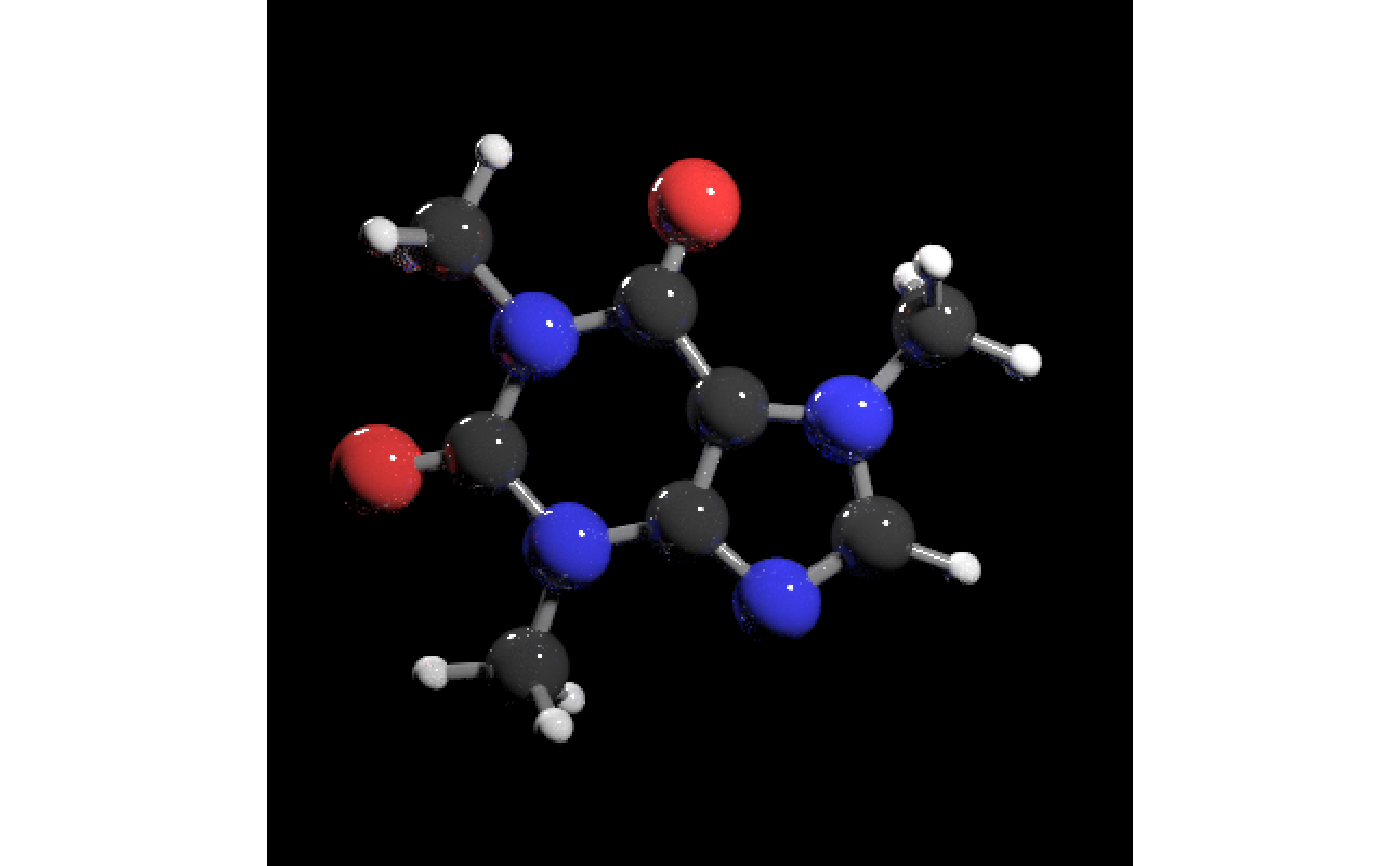
# Generate a scene with caffeine molecule with just the atoms
# \donttest{
get_example_molecule("caffeine") |>
read_sdf() |>
generate_full_scene() |>
render_model(samples=256,sample_method="sobol_blue")
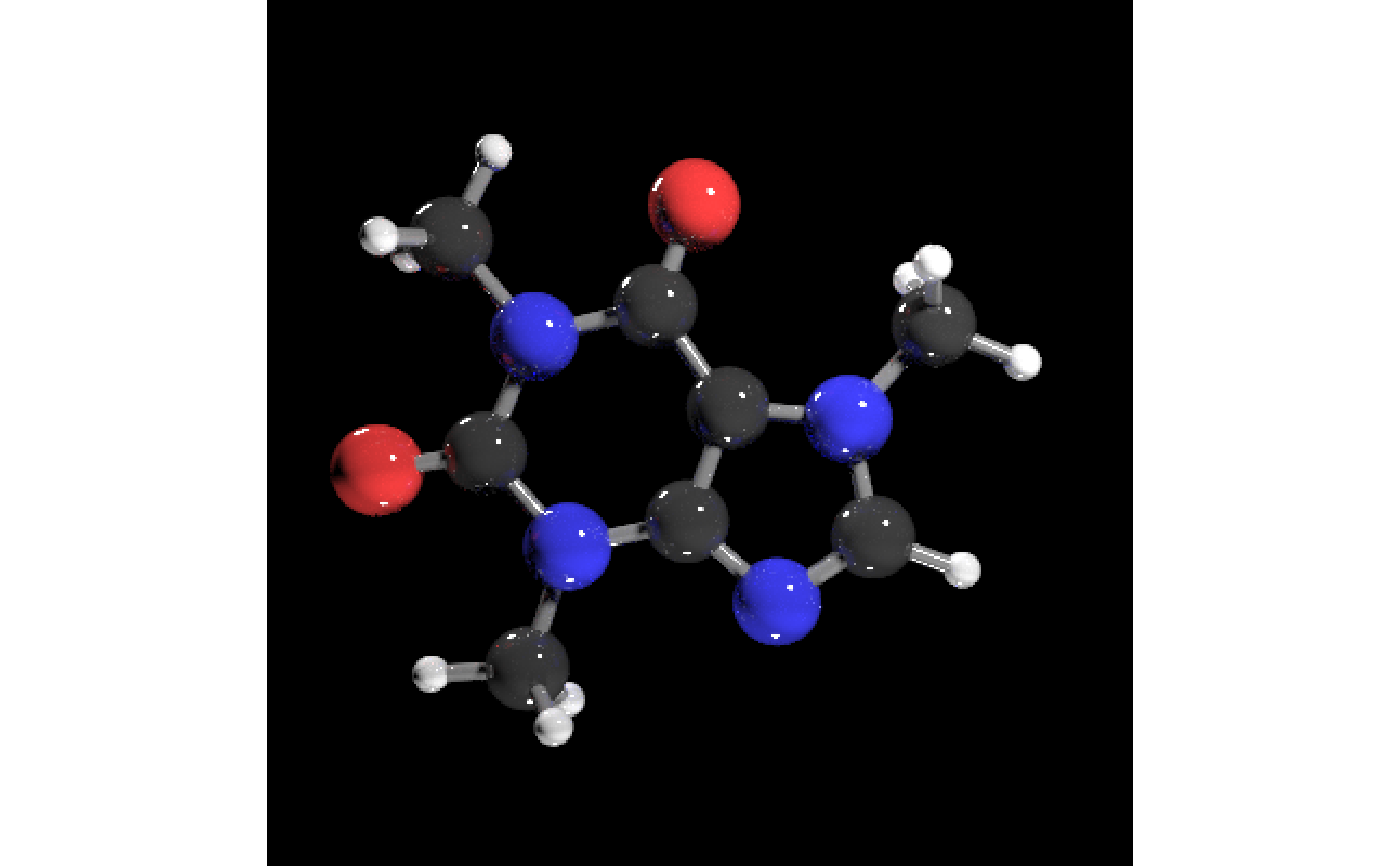
 #Light the example from below as well
get_example_molecule("caffeine") |>
read_sdf() |>
generate_full_scene() |>
render_model(lights = "both", samples=256,sample_method="sobol_blue")
#Light the example from below as well
get_example_molecule("caffeine") |>
read_sdf() |>
generate_full_scene() |>
render_model(lights = "both", samples=256,sample_method="sobol_blue")
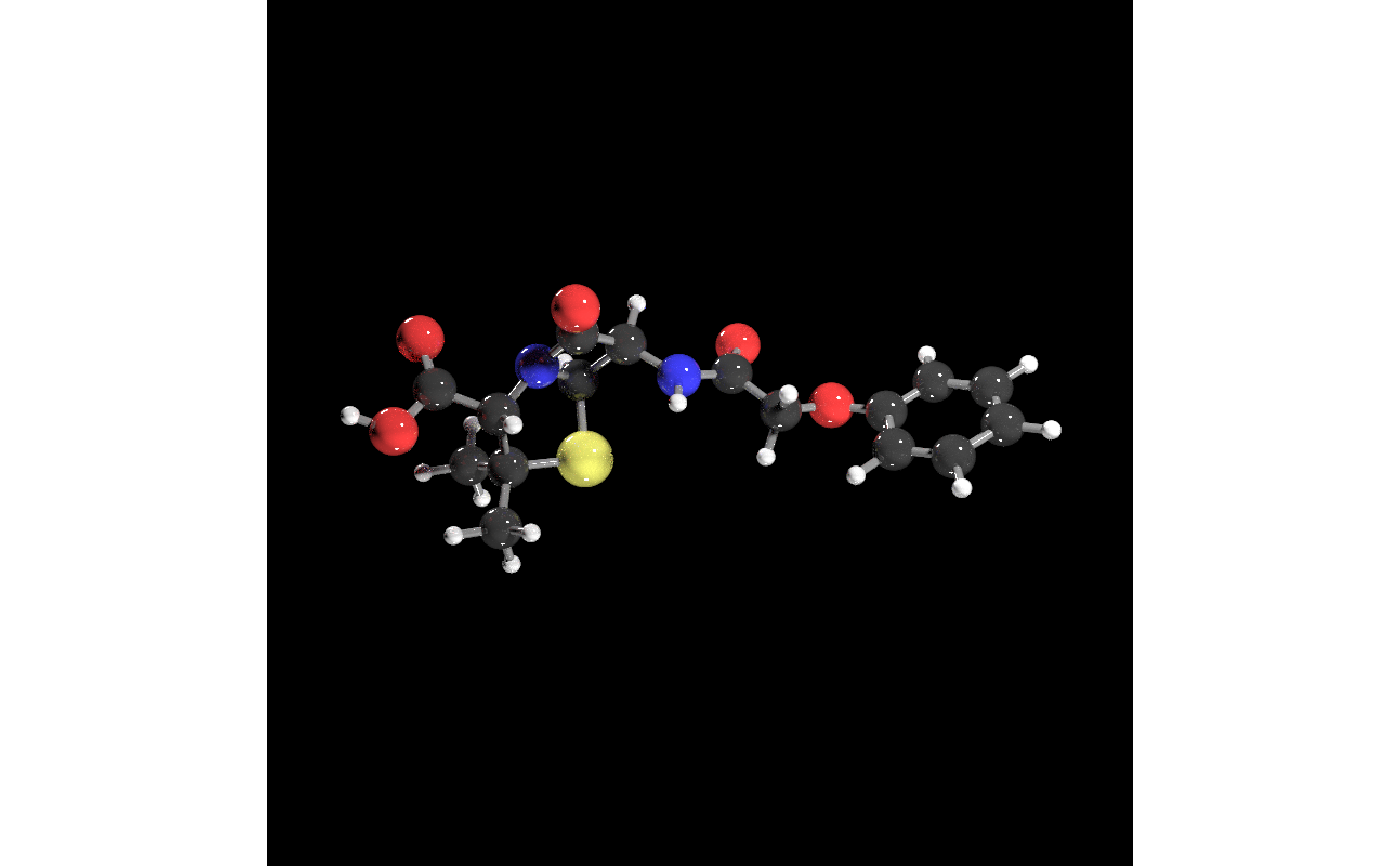
 #Generate a scene with penicillin, increasing the number of samples and the width/height
#for a higher quality render.
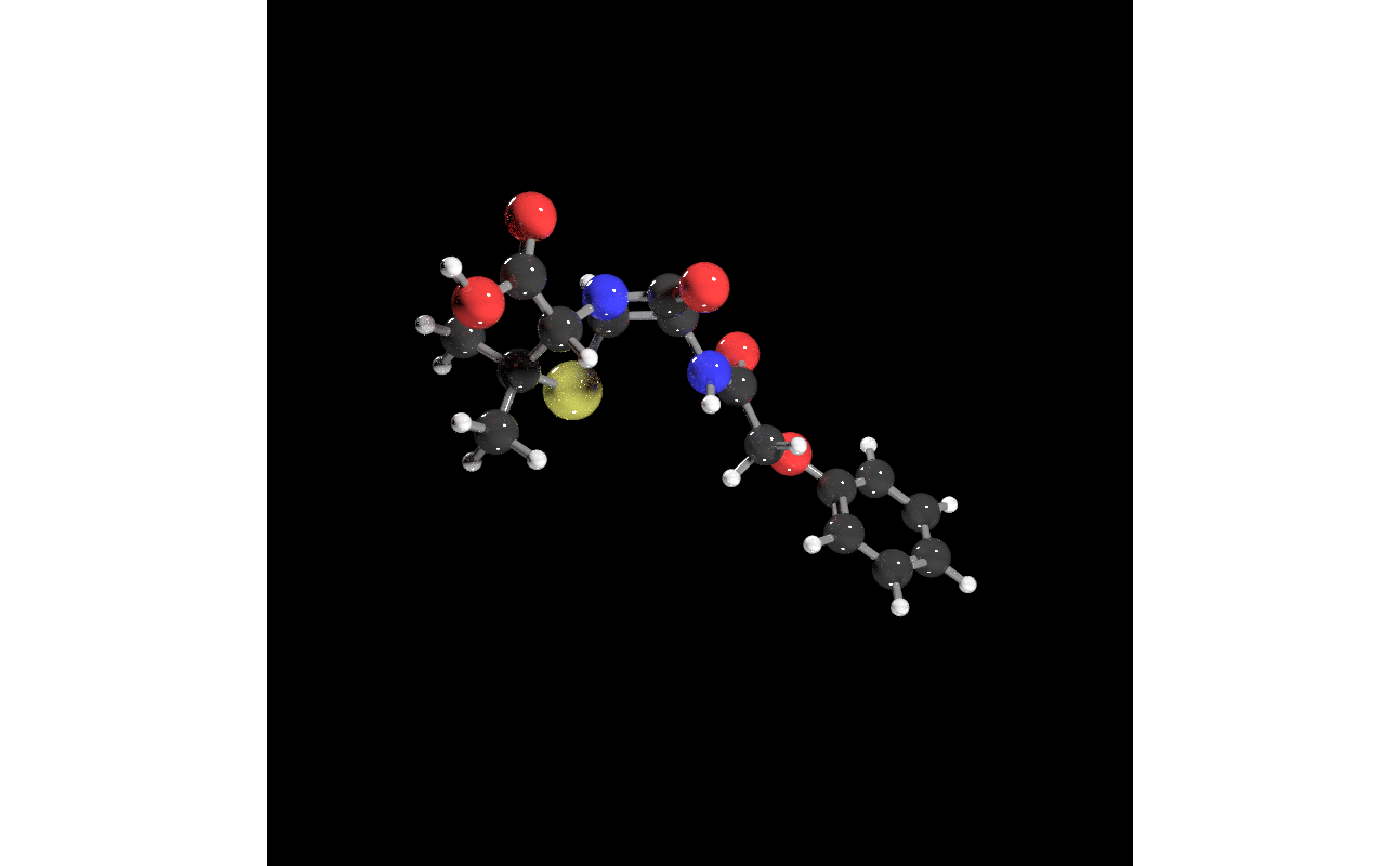
get_example_molecule("penicillin") |>
read_sdf() |>
generate_full_scene() |>
render_model(lights = "both", samples=256, width=800, height=800,sample_method="sobol_blue")
#Generate a scene with penicillin, increasing the number of samples and the width/height
#for a higher quality render.
get_example_molecule("penicillin") |>
read_sdf() |>
generate_full_scene() |>
render_model(lights = "both", samples=256, width=800, height=800,sample_method="sobol_blue")
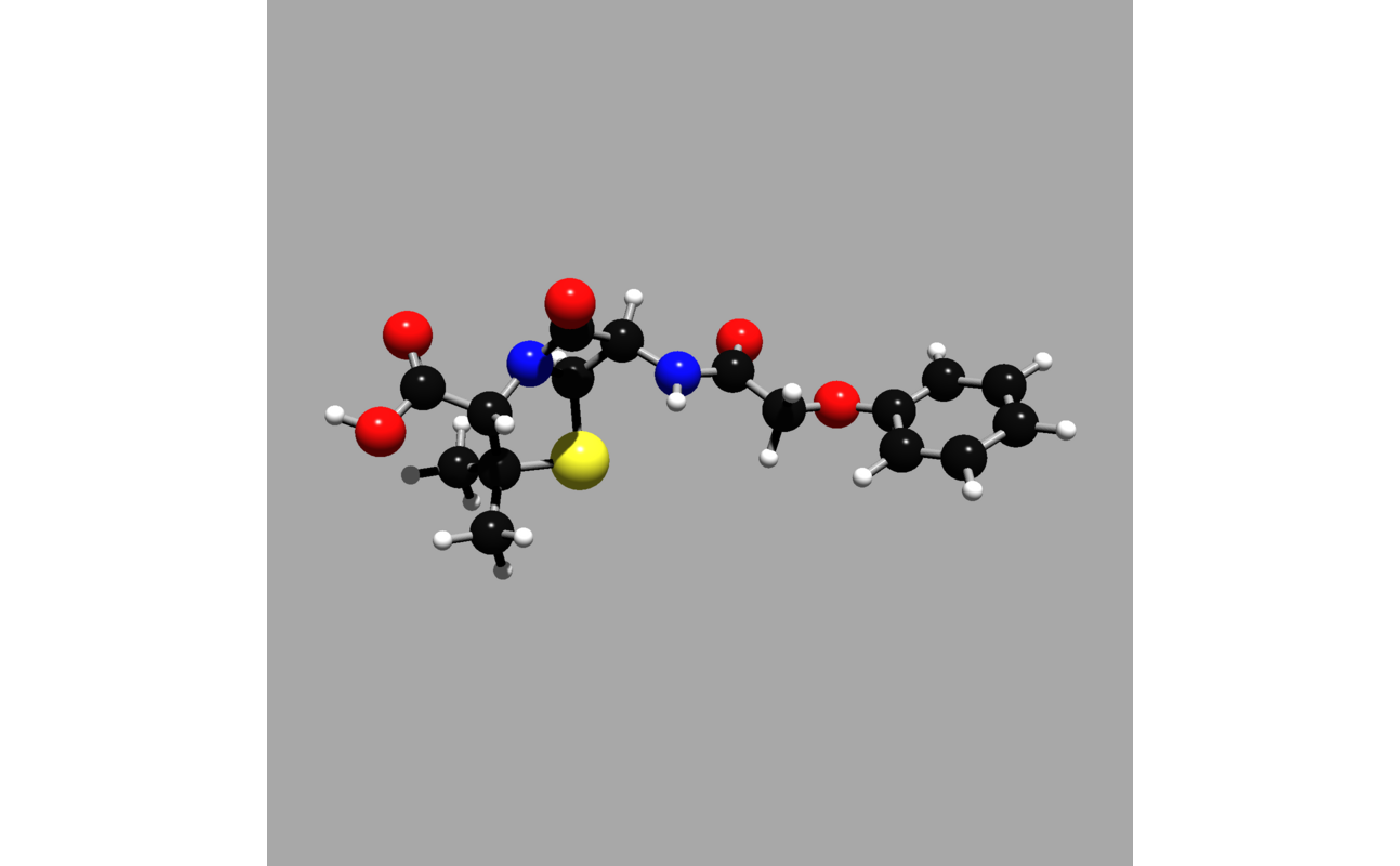
 #Render the scene with rayvertex and custom lights
get_example_molecule("penicillin") |>
read_sdf() |>
generate_full_scene(pathtrace=FALSE) |>
render_model(width=800, height=800,background="grey66",
lights = rayvertex::directional_light(c(0.2,1,1)))
#Render the scene with rayvertex and custom lights
get_example_molecule("penicillin") |>
read_sdf() |>
generate_full_scene(pathtrace=FALSE) |>
render_model(width=800, height=800,background="grey66",
lights = rayvertex::directional_light(c(0.2,1,1)))
 #Rotate the molecule 30 degrees around the y axis, and the 30 degrees around the z axis
get_example_molecule("penicillin") |>
read_sdf() |>
generate_full_scene() |>
render_model(lights = "both", samples=256, width=800, height=800,
angle=c(0,30,30),sample_method="sobol_blue")
#Rotate the molecule 30 degrees around the y axis, and the 30 degrees around the z axis
get_example_molecule("penicillin") |>
read_sdf() |>
generate_full_scene() |>
render_model(lights = "both", samples=256, width=800, height=800,
angle=c(0,30,30),sample_method="sobol_blue")
 #Add a checkered plane underneath, using rayrender::add_object and rayrender::xz_rect().
#We also pass a value to `clamp_value` to minimize fireflies (bright spots).
library(rayrender)
#>
#> Attaching package: ‘rayrender’
#> The following object is masked from ‘package:raymolecule’:
#>
#> run_documentation
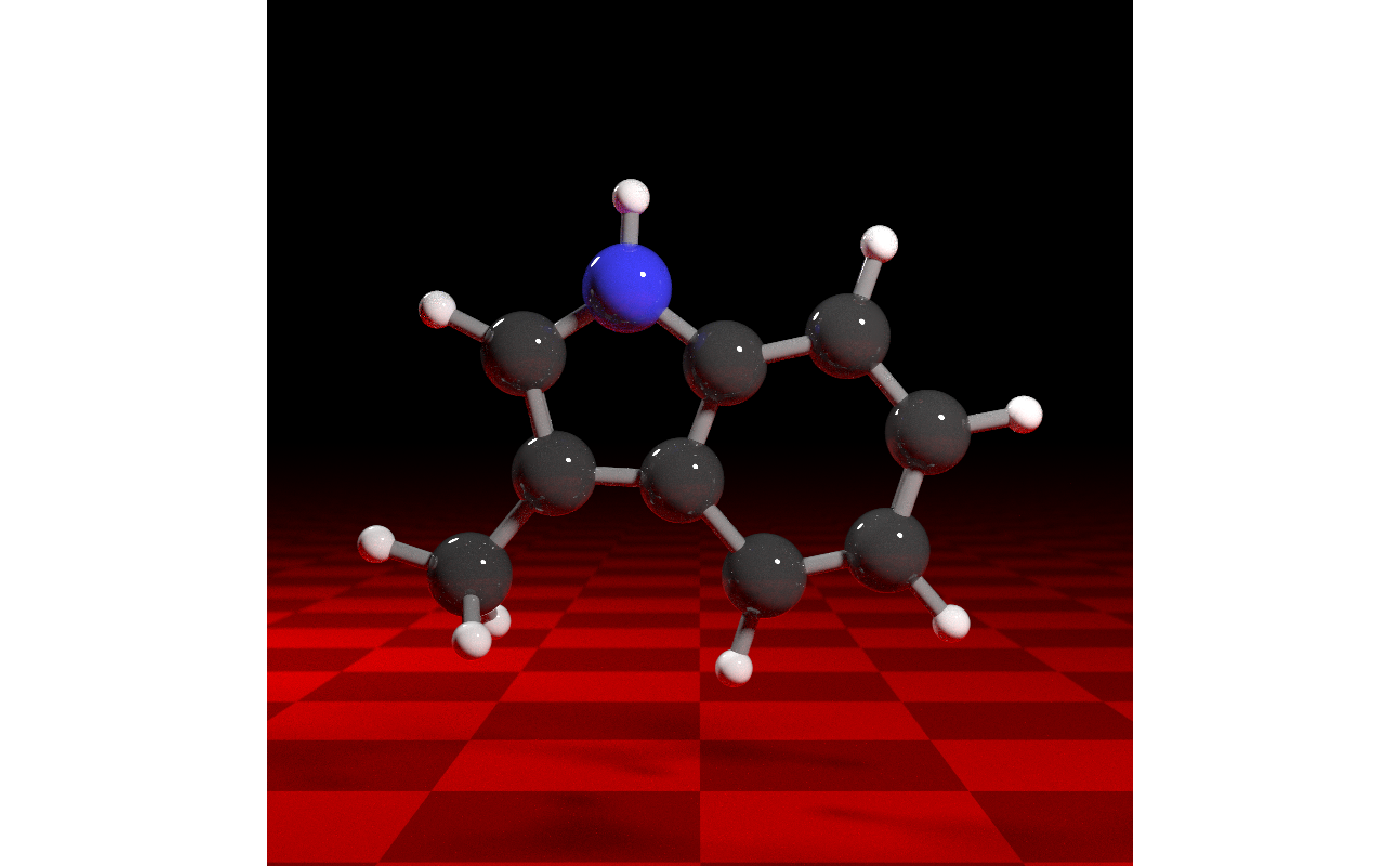
get_example_molecule("skatole") |>
read_sdf() |>
generate_full_scene() |>
add_object(xz_rect(xwidth=1000,zwidth=1000,y=-4,
material=diffuse(color="#330000",checkercolor="#770000"))) |>
render_model(samples=256, width=800, height=800, clamp_value=10,
sample_method="sobol_blue")
#Add a checkered plane underneath, using rayrender::add_object and rayrender::xz_rect().
#We also pass a value to `clamp_value` to minimize fireflies (bright spots).
library(rayrender)
#>
#> Attaching package: ‘rayrender’
#> The following object is masked from ‘package:raymolecule’:
#>
#> run_documentation
get_example_molecule("skatole") |>
read_sdf() |>
generate_full_scene() |>
add_object(xz_rect(xwidth=1000,zwidth=1000,y=-4,
material=diffuse(color="#330000",checkercolor="#770000"))) |>
render_model(samples=256, width=800, height=800, clamp_value=10,
sample_method="sobol_blue")
 # }
# }